Jonathan (Jack) Goetz
Jack’s research is motivated by problems in computational chemistry, largely aimed at reducing the number of simulations required to understand a modeled chemical reaction. One approach is using active learning. Active learning algorithms use both unlabelled and available labelled data to sequentially select data points to label, with the goal of building the best possible model with a limited budget of labelled data. Jack developed new algorithms for active learning in non-parametric regression to identify the most useful simulations, allowing others to be safely skipped.
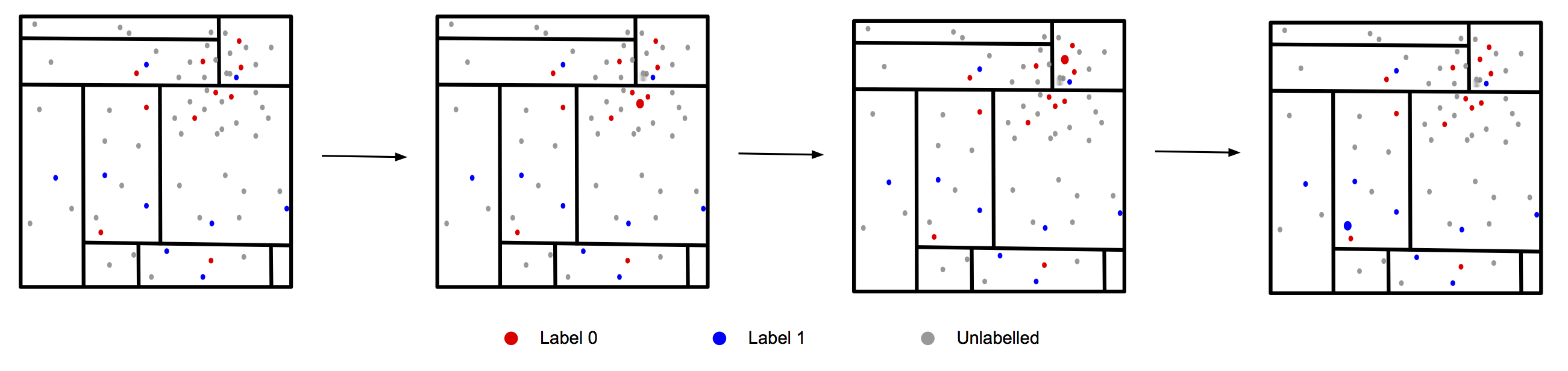
The figure below shows an active learning example using a Mondrian Tree: With a small amount of labelled data the active learning algorithm picks the next unlabelled data point, selecting the point it believes is most informative. Once it gets the label it reassess the informativeness of each point before selecting the next one.
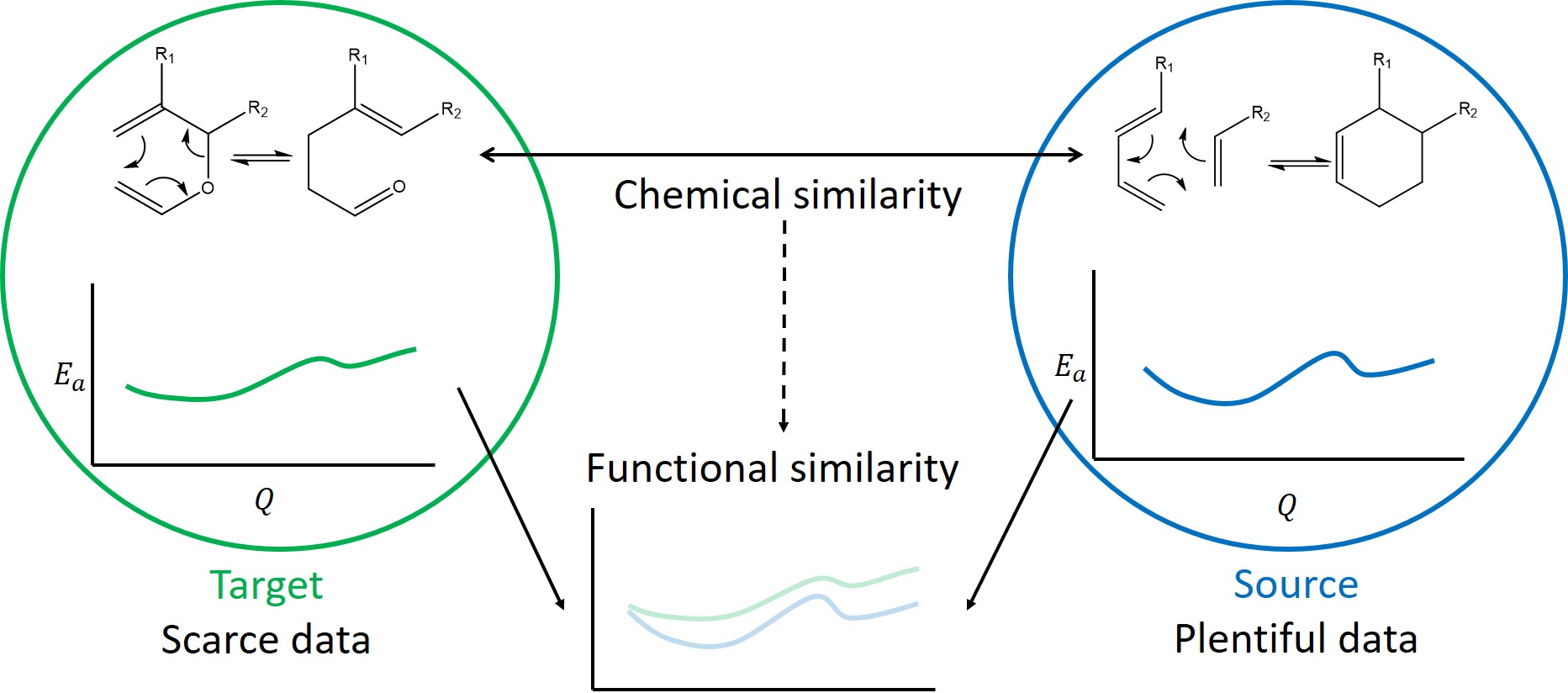
Another useful tool is transfer learning, which aims to reuse labelled data from one distribution (called the source) to aid in prediction on a different, but related distribution (called the target). By specializing these methods for computational chemistry data used to make predictions on one family of molecules can be reused to assist in prediction on chemically similar families of molecules.
The figure below shows a transfer learning example: The energy profiles of these two families of reactions are similar due to their shared chemical structure, but not identical.
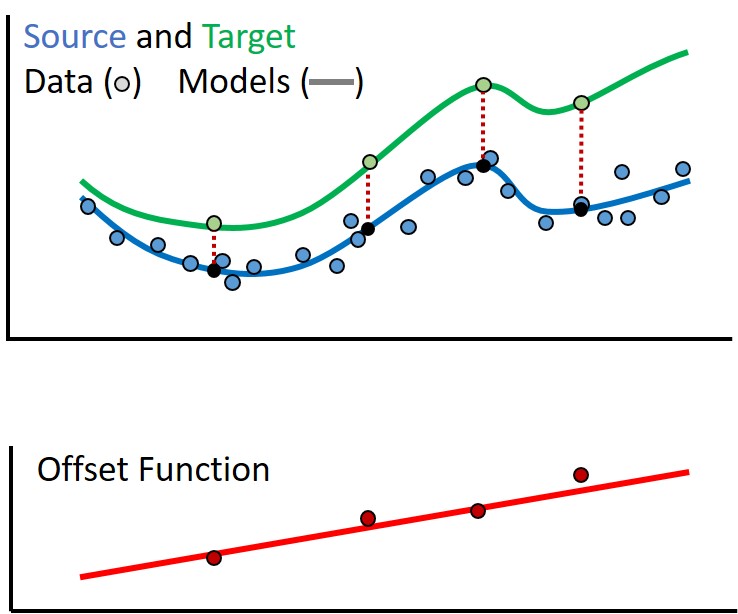
However with a few data points from the target, we can model the offset between the energy profiles as shown in the figure below. By adding the model for the offset (red) to the model for the source (blue), we create a good model for the target (green). Thanks to Alan Chien for help with these graphics.